Molecule Generation with Fragment Retrieval Augmentation
Conference on Neural Information Processing Systems (NeurIPS), 2024
Seul Lee1*
Karsten Kreis2
Srimukh Prasad Veccham2
Meng Liu2
Danny Reidenbach2
Saee Paliwal2
Arash Vahdat2†
Weili Nie2†
1
2
* Work done during an internship at NVIDIA
† Equal advising
seul.lee@kaist.ac.kr
{kkreis, sveccham, menliu, dreidenbach, saeep, avahdat, wnie}@nvidia.com
[Paper]
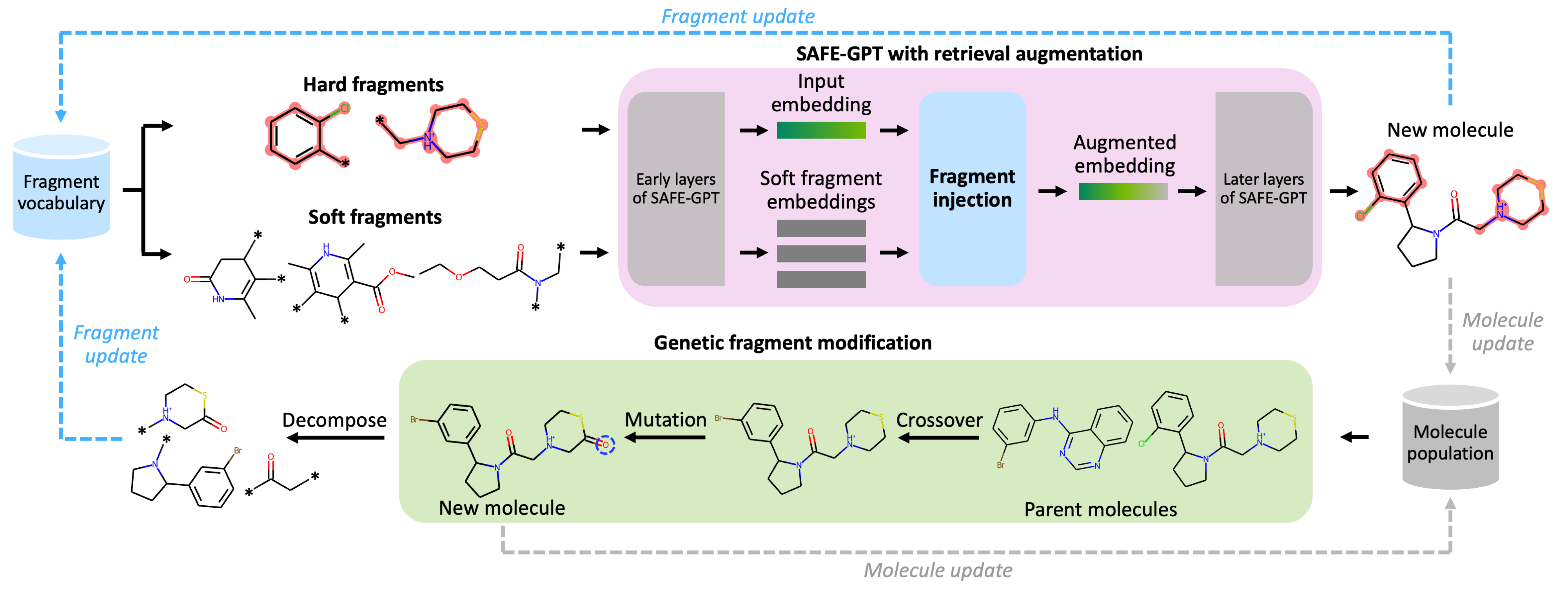
f-RAG: Fragment Retrieval-Augmented Generation
Many fragment-based molecule generation methods show limited exploration beyond the existing fragments in the database as they only reassemble or slightly modify the given ones. To tackle this problem, we propose a new fragment-based molecule generation framework with retrieval augmentation, namely Fragment Retrieval-Augmented Generation (f-RAG). f-RAG is based on a pre-trained molecular generative model (SAFE-GPT) that proposes additional fragments from input fragments to complete and generate a new molecule. Given a fragment vocabulary, f-RAG retrieves two types of fragments: (1) hard fragments, which serve as building blocks that will be explicitly included in the newly generated molecule, and (2) soft fragments, which serve as reference to guide the generation of new fragments through a trainable fragment injection module. To extrapolate beyond the existing fragments, f-RAG updates the fragment vocabulary with generated fragments via an iterative refinement process which is further enhanced with post-hoc genetic fragment modification. f-RAG can achieve an improved exploration-exploitation trade-off by maintaining a pool of fragments and expanding it with novel and high-quality fragments through a strong generative prior.
Fragment-based Drug Discovery with f-RAG
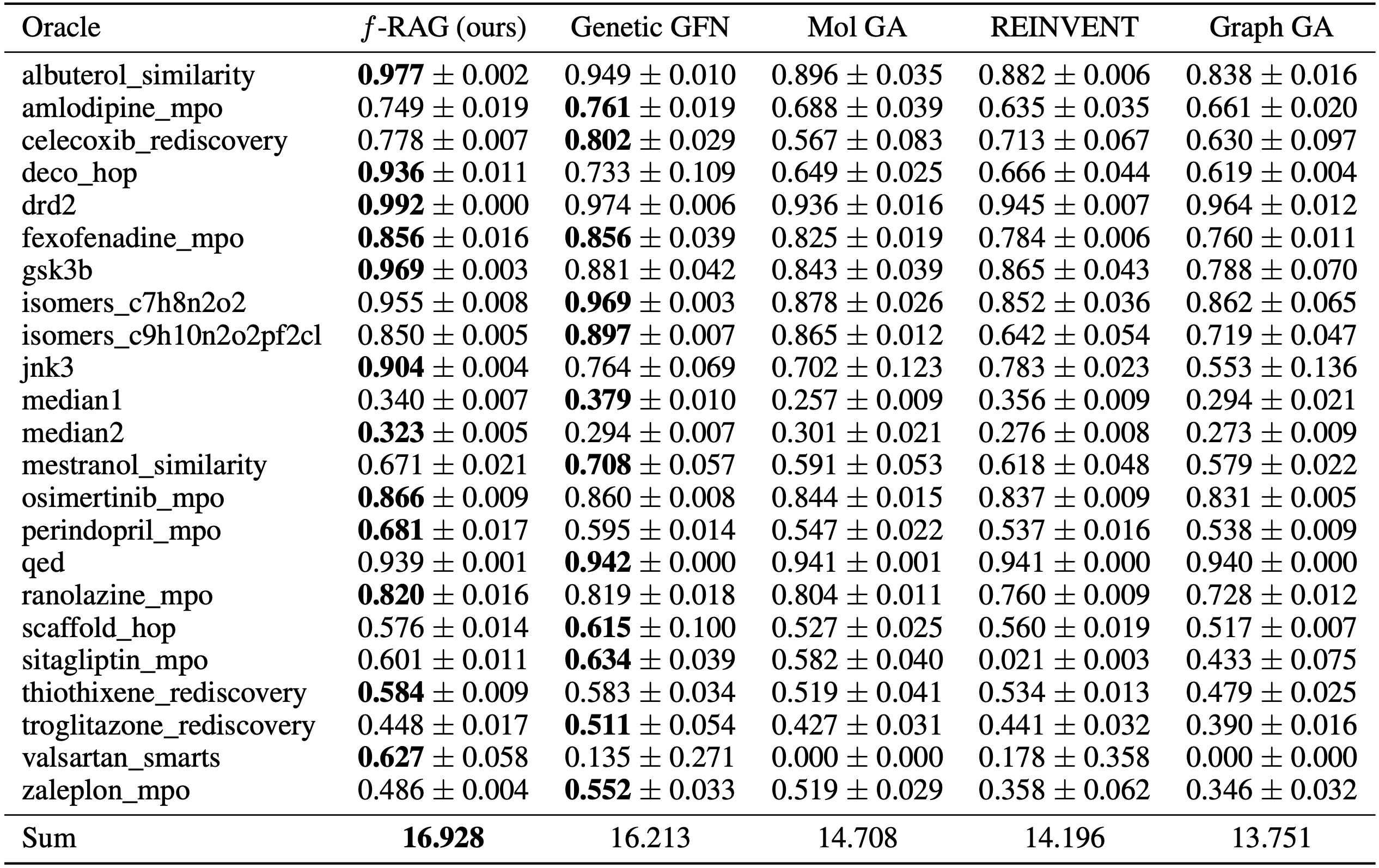
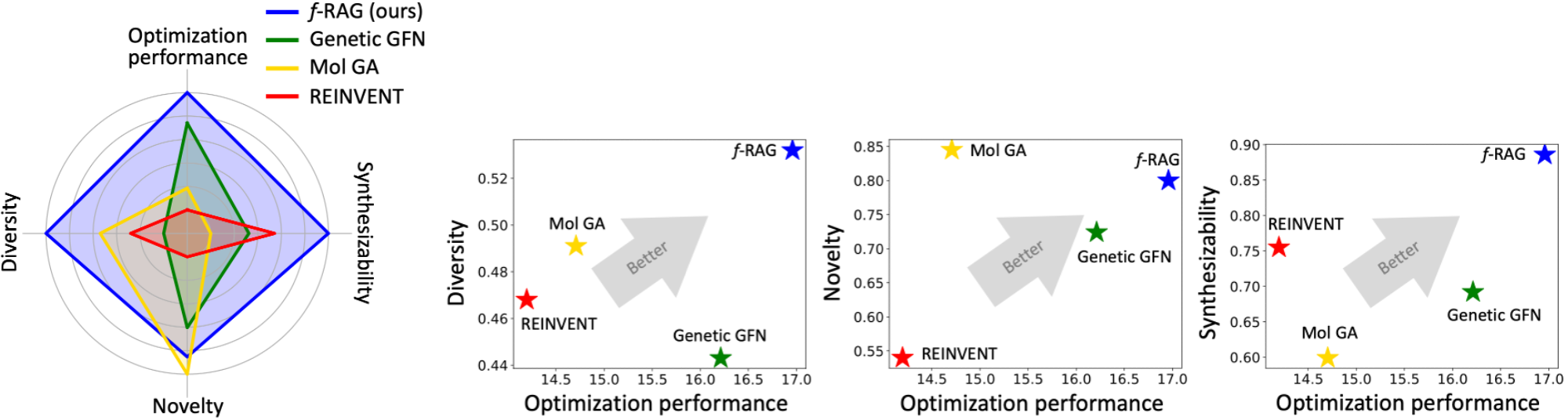
On the PMO benchmark, f-RAG outperforms the previous methods in terms of the sum of the AUC top-10 values and achieves the highest AUC top-10 values in 12 out of 23 tasks. Furthermore, even though the essential considerations in drug discovery (e.g., diversity, novelty, and synthesizability) often conflict with each other, f-RAG exhibits the best balance across them, demonstrating its applicability as a promising tool for drug discovery.
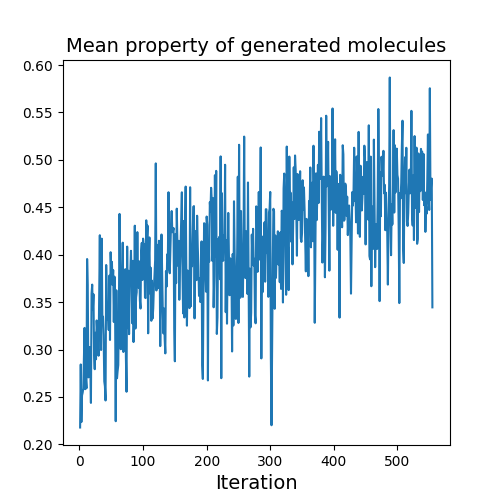
Evolution of Generated Molecules over Iterations
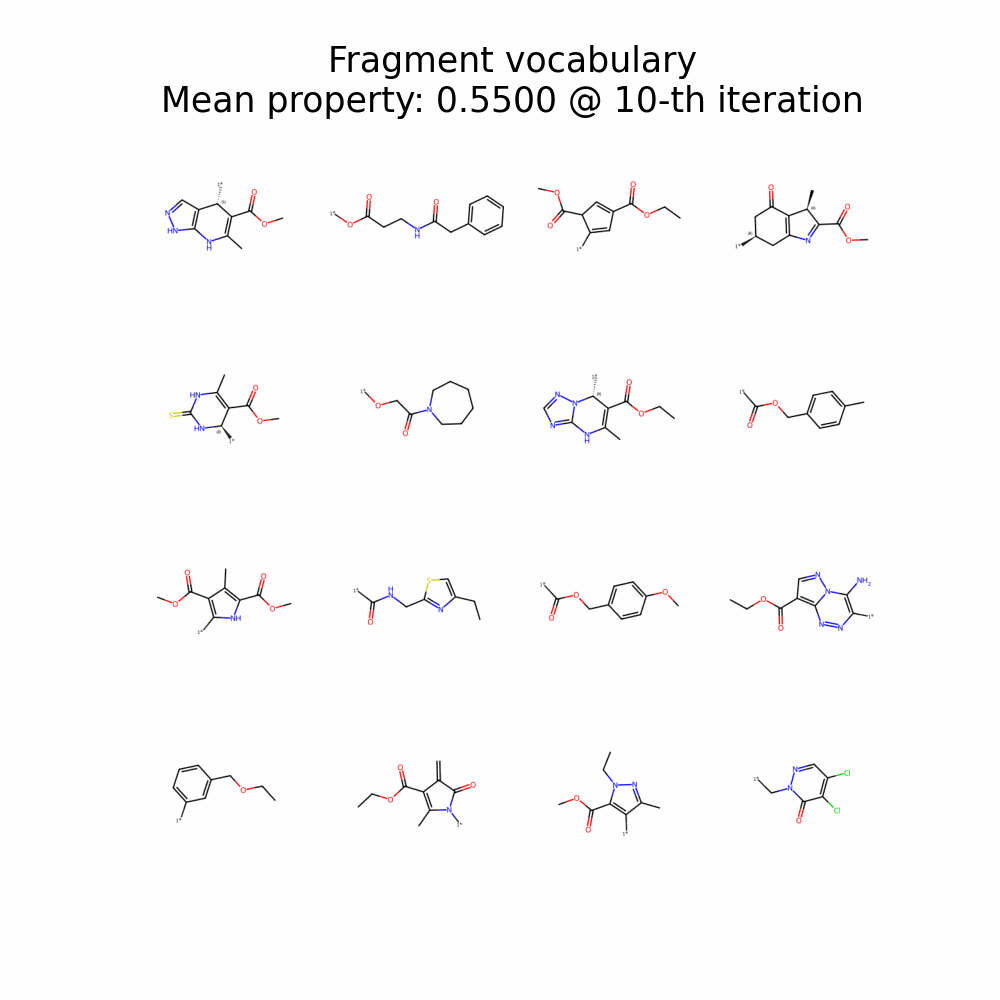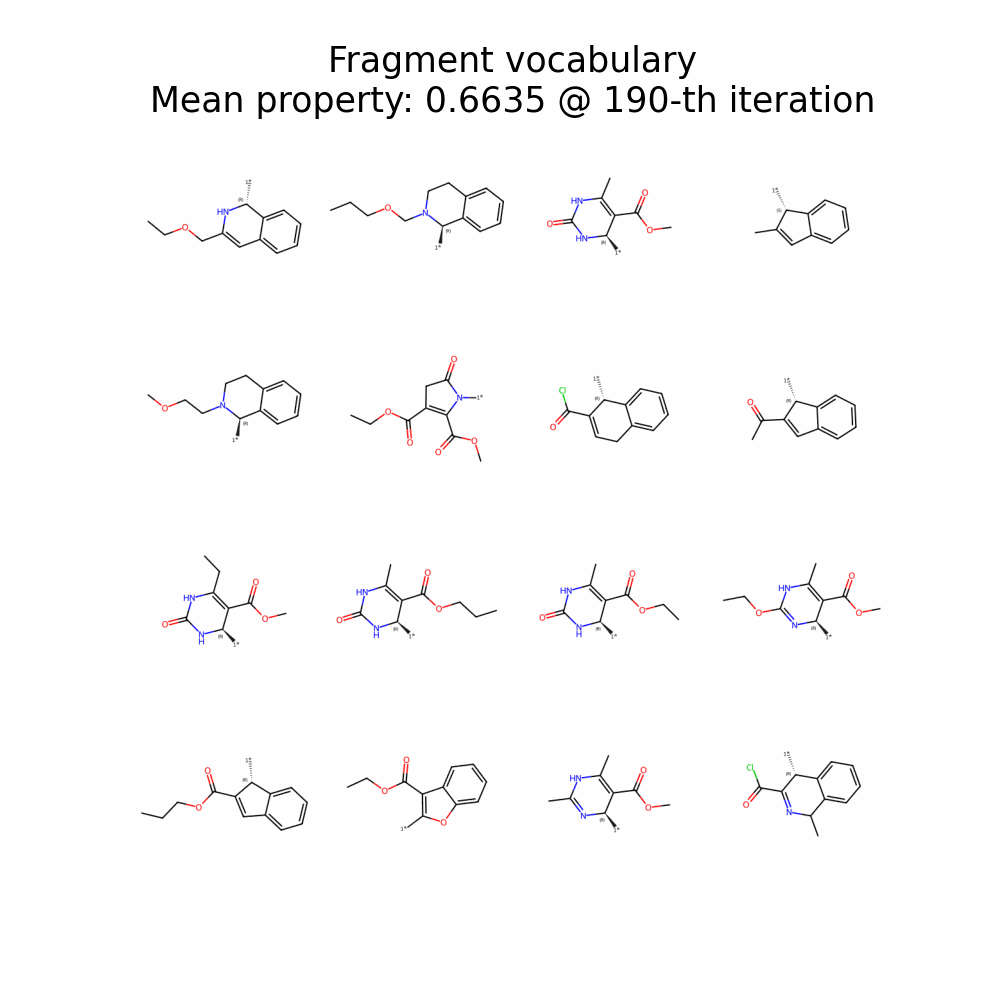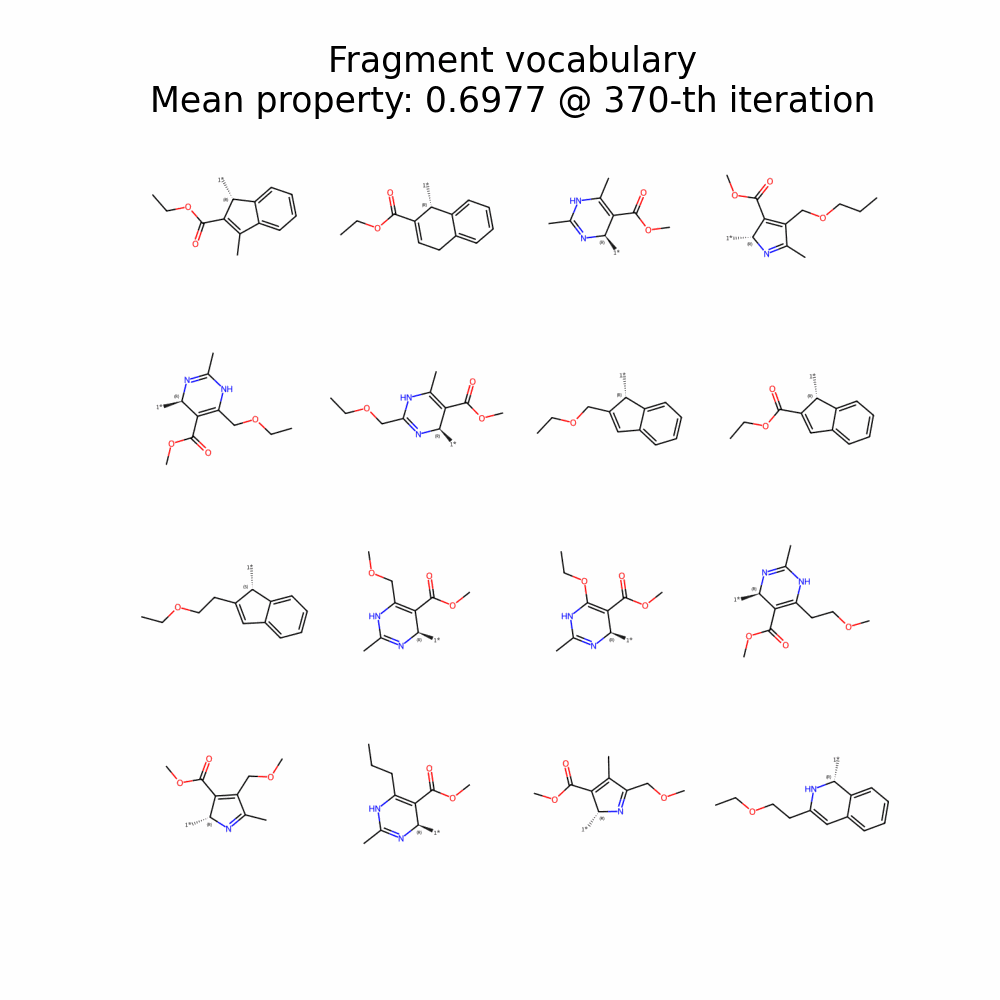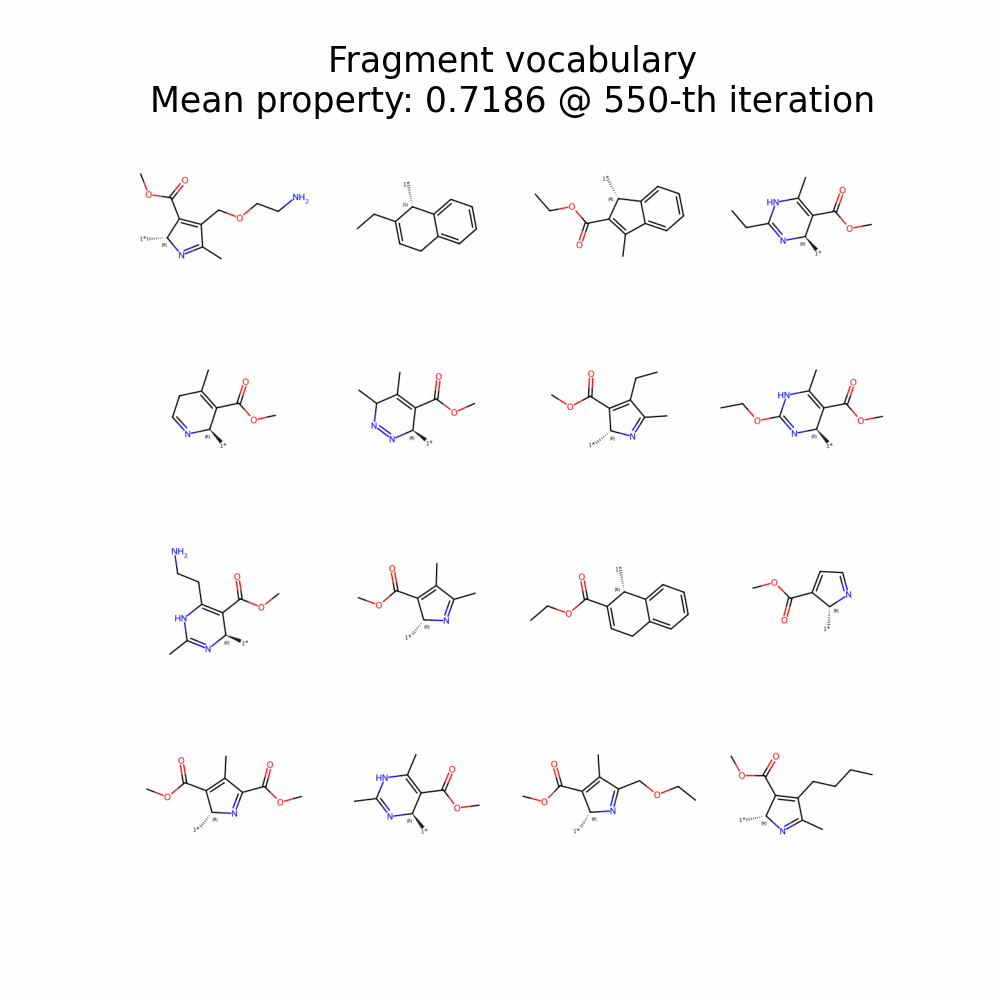
f-RAG dynamically updates the fragment vocabulary with newly generated fragments. The fragment vocabulary and therefore the generated molecules are iteratively refined throughout generation.
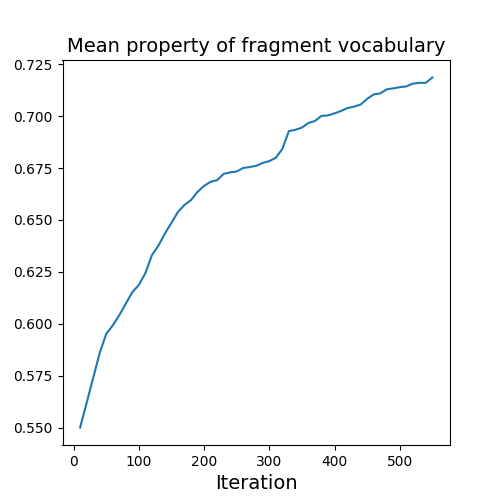
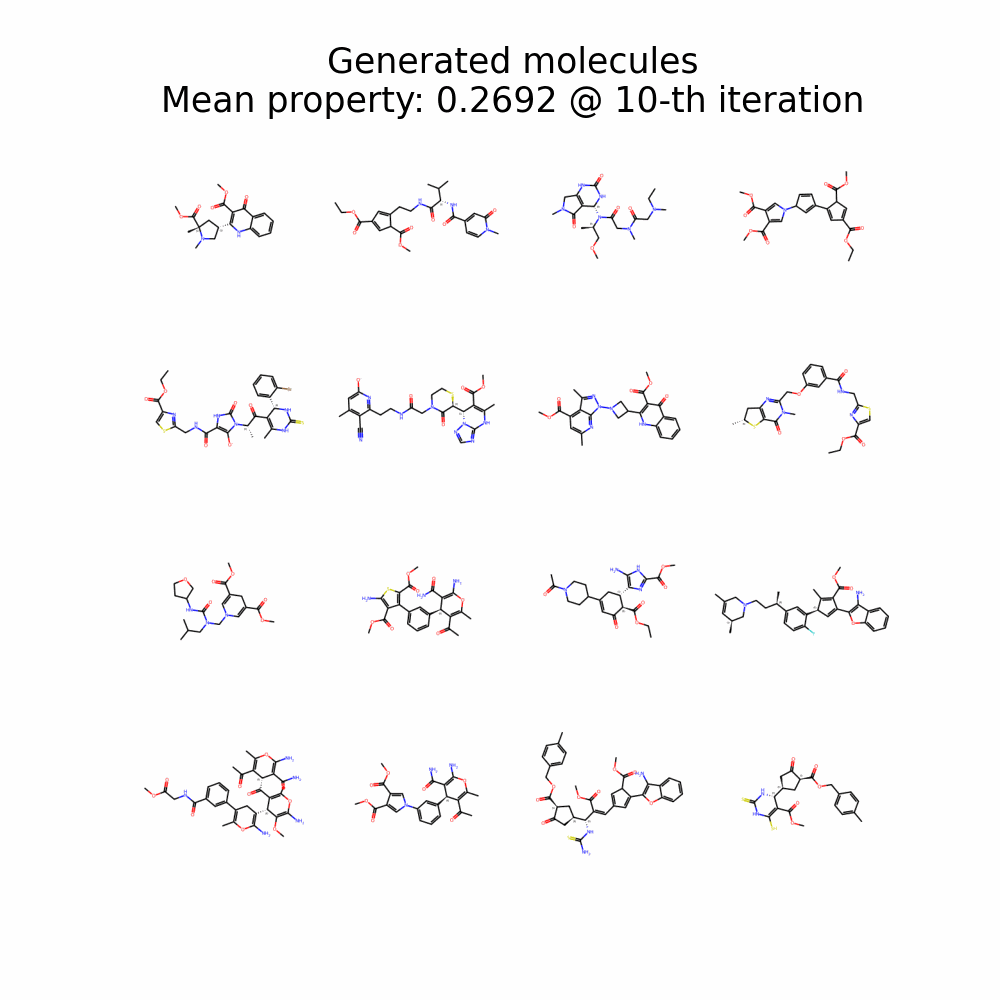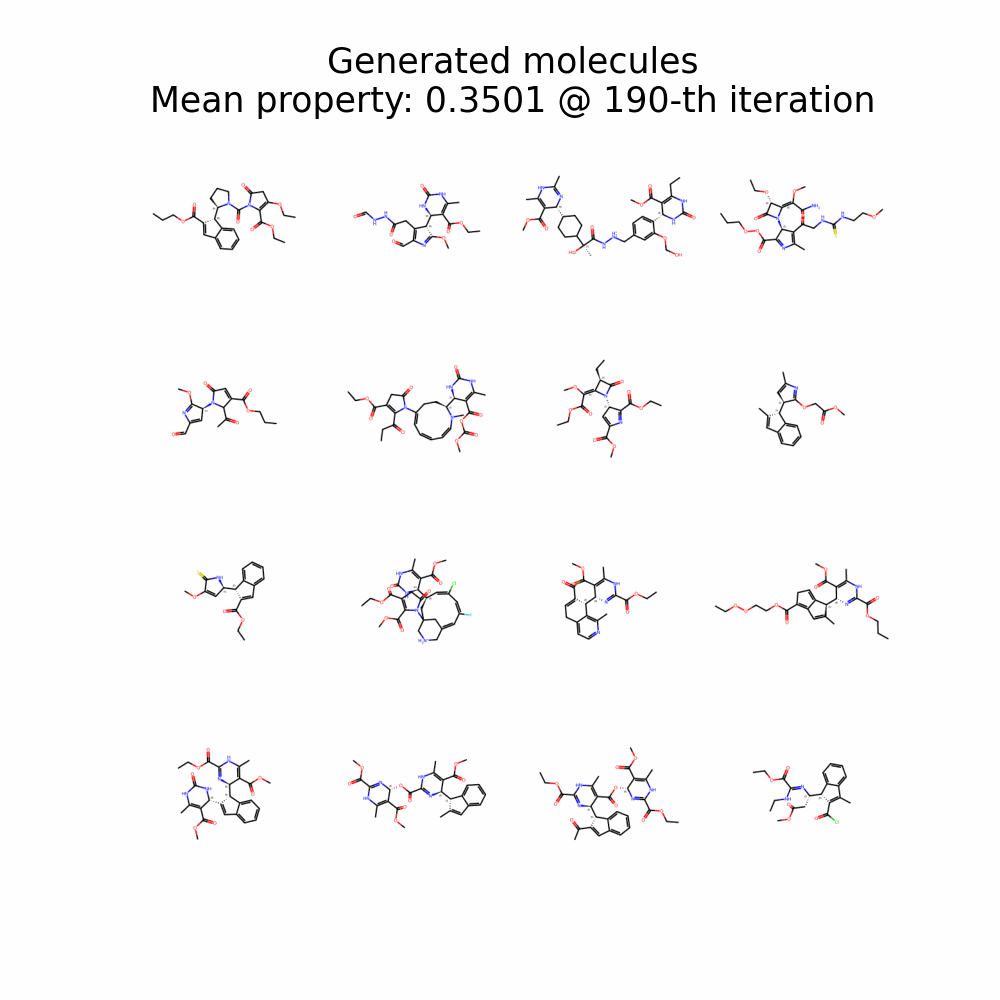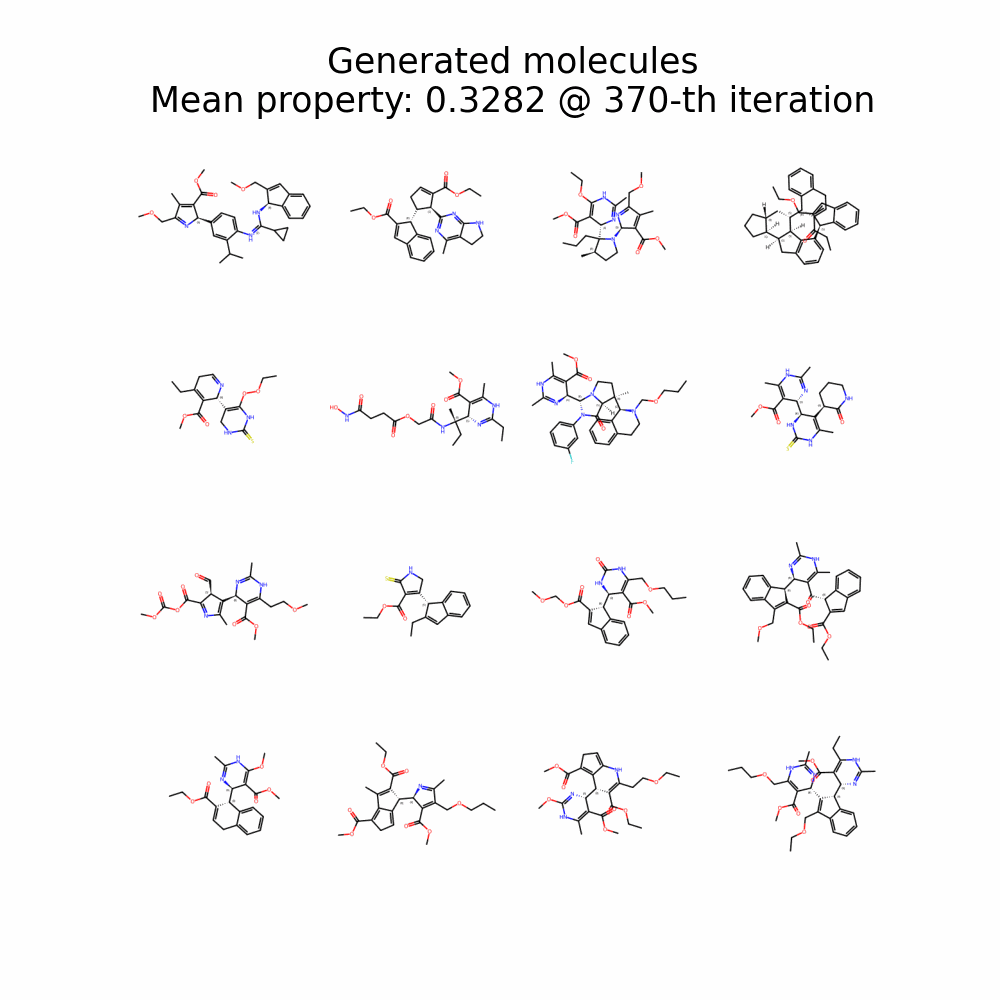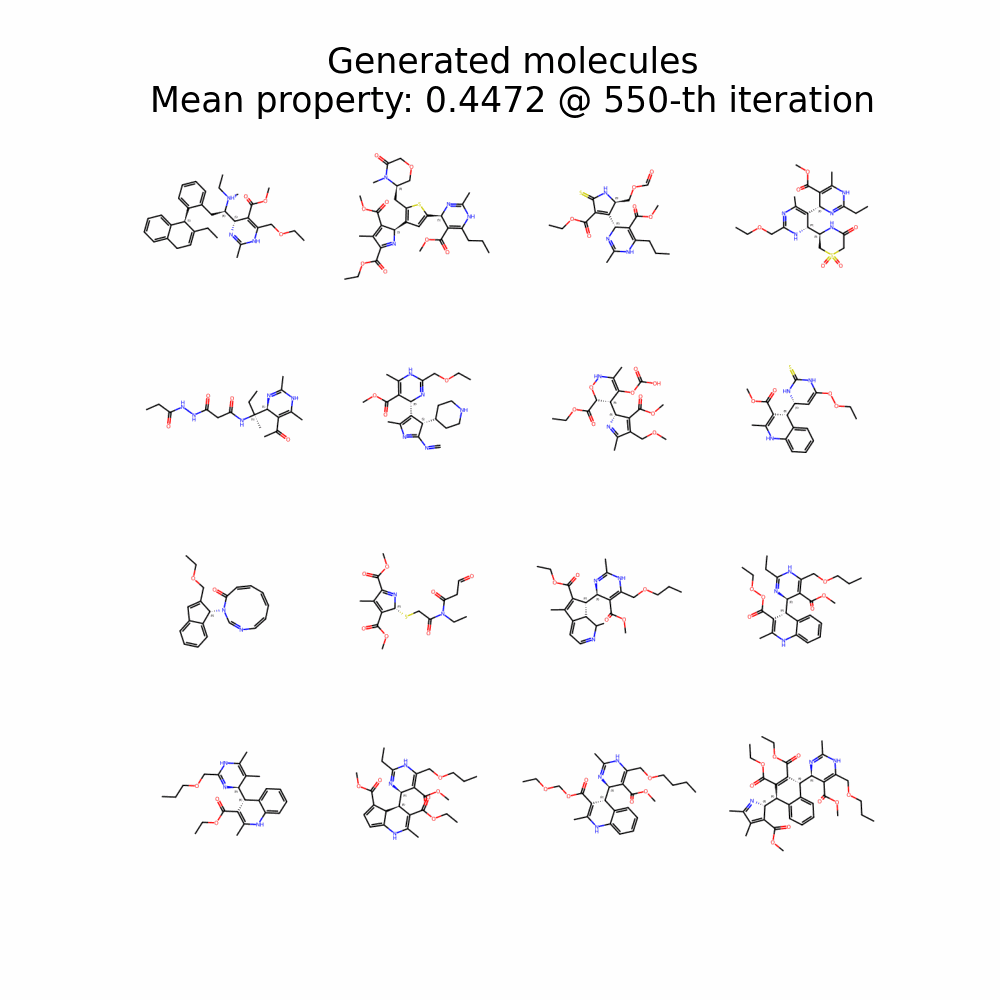
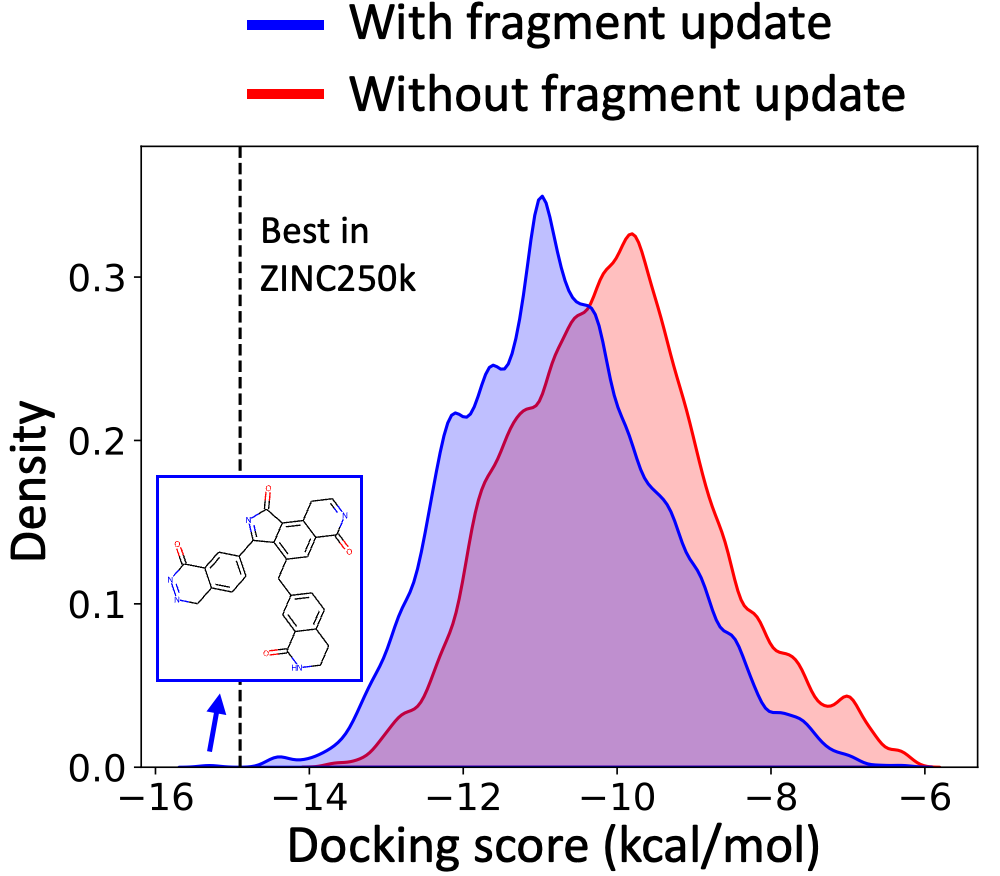
Exploration with Dynamic Vocabulary Update
f-RAG effectively improves the exploration-exploitation trade-off in drug discovery by utilizing existing fragments while dynamically updating the fragment vocabulary with newly proposed fragments. f-RAG can discover molecules that have better target properties than the top molecule in the training set with the dynamic update.
Citation
@article{lee2024frag,
title = {Molecule Generation with Fragment Retrieval Augmentation},
author = {Seul Lee and Karsten Kreis and Srimukh Prasad Veccham and Meng Liu and
Danny Reidenbach and Saee Paliwal and Arash Vahdat and Weili Nie},
journal = {Advances in Neural Information Processing Systems},
year = {2024}
}